Glossary¶
- TCR
T-cell receptor. A TCR consists of one α and one β chain (or, alternatively, one γ and one δ chain). Each chain consists of a constant and a variable region. The variable region is responsible for antigen recognition, mediated by CDR regions.
Scirpy currently only supports α/β-TCRs. For more information, see the page about our TCR model.
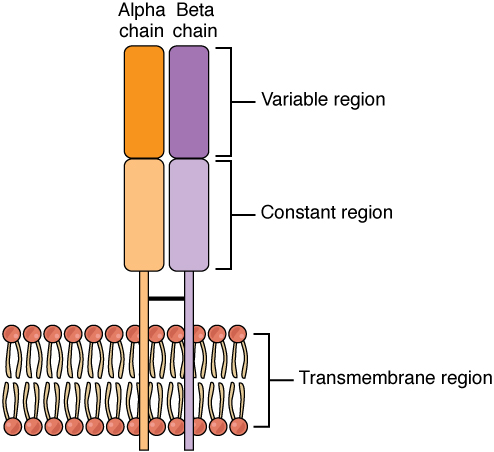
Image from Wikimedia commons under the CC BY-3.0 license.¶
- Clonotype
A clonotype designates a collection of T or B cells that descend from a common, antecedent cell, and therefore, bear the same adaptive immune receptors and recognize the same epitopes.
In single-cell RNA-sequencing (scRNA-seq) data, T cells sharing identical complementarity-determining regions 3 (CDR3) nucleotide sequences of both α and β TCR chains make up a clonotype.
Scirpy provides a flexible approach to clonotype definition based on CDR3 sequence identity or similarity. Additionally, it is possible to require clonotypes to have the same V-gene, enforcing the CDR 1 and 2 regions to be the same.
For more details, see the page about our TCR model and the API documentation of
scirpy.tl.define_clonotypes().- Clonotype cluster
A higher-order aggregation of clonotypes that have different CDR3 nucleotide sequences, but might recognize the same antigen because they have the same or similar CDR3 amino acid sequence.
See also:
scirpy.tl.define_clonotype_clusters().- Private clonotype
A clonotype that is specific for a certain patient.
- Public clonotype
A clonotype that is shared across multiple patients, e.g. a clonotype recognizing common viral epitope.
- Tissue-specific clonotype
A clonotype that only occurs in a certain tissue of a certain patient.
- Multi-tissue clonotype
A clonotype that occurs in multiple tissues of the same patient.
- Convergent evolution of clonotypes
It has been proposed that TCRs are subject to convergent evolution, i.e. a selection pressure that leads to TCRs recognizing the same antigen ([VKP+06]).
Evidence of convergent evolution could be clonotypes with the same CDR3 amino acid sequence, but different CDR3 nucleotide sequences (due to synonymous codons) or clonotypes with highly similar CDR3 amino acid sequences that recognize the same antigen.
- Epitope
The part of an antigen that is recognized by the TCR (or B-cell receptor, or antibody).
- CDR3
Complementary-determining region 3. See CDR.
- CDR
Complementary-determining region. The diversity and, therefore, antigen-specificity of TCRs is predominanly determined by three hypervariable loops (CDR1, CDR2, and CDR3) on each of the α- and β receptor arms.
CDR1 and CDR2 are fully encoded in germline V genes. In contrast, the CDR3 loops are assembled from V and J segments (TCR-α) and V, D and J segments (TCR-β) and comprise random additions and deletions at the junction sites (see also V(D)J). Thus, CDR3 regions make up a large part of the TCR variability and are therefore thought to be particularly important for antigen specificity (reviewed in [AHS15]).
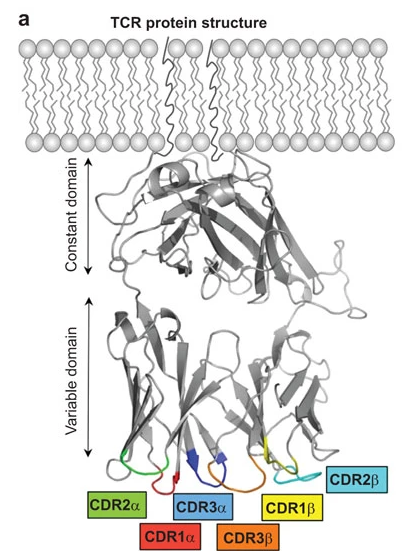
Image from [AHS15] under the CC BY-NC-SA-3.0 license.¶
- V(D)J
The variability of TCR chain sequences originates from the genetic recombination of Variable, Diversity and Joining gene segments. The TCR-α chain gets assembled from V and J loci only, the TCR-β chain from all three V, D and J loci.
As an example, the figure below shows how a TCR-α chain is assembed from the tra locus. V to J recombination joins one of many TRAV segments to one of many TRAJ segments. Next, introns are spliced out, resulting in a TCR-α chain transcript with V, J and C segments directly next to each other (reviewed in [AHS15]).
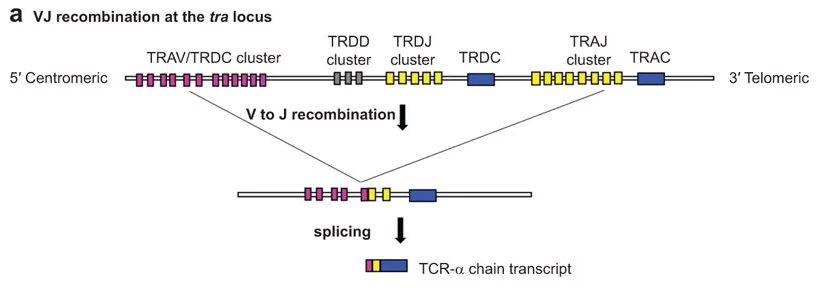
Image from [AHS15] under the CC BY-NC-SA-3.0 license.¶
- Dual TCR
TCRs with more than one pair of α- and β chains. While this was previously thought to be impossible due to the mechanism of allelic exclusion ([BSB10]), there is an increasing amound of evidence for a bona fide dual-TCR population ([SB19], [JPG10]).
For more information on how Scirpy handles dual TCRs, see the page about our TCR model.
- Multichain-cell
Cells with more than two α- and β chains that do not fit into the Dual TCR model. These are usually rare and could be explained by doublets/multiplets, i.e. two ore more cells that were captured in the same droplet.
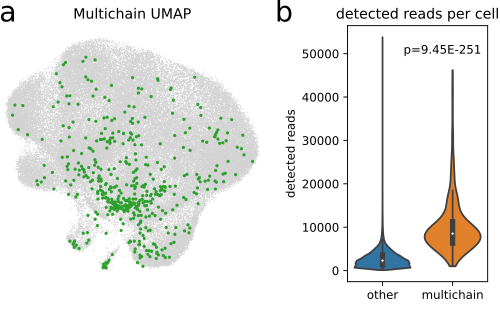
(a) UMAP plot of 96,000 cells from [WMdA+20] with at least one detected CDR3 sequence with multichain-cells (n=474) highlighted in green. (b) Comparison of detected reads per cell in multichain-cells and other cells. Multichain cells comprised significantly more reads per cell (p = 9.45 × 10−251, Wilcoxon-Mann-Whitney-test), supporting the hypothesis that (most of) multichain cells are technical artifacts arising from cell-multiplets ([IKK+16]).¶
- Orphan chain
A TCR chain is called orphan, if its corresponding counterpart has not been detected. For instance, if a cell has only a TCR-α chain, but no TCR-β chain, the cell will be flagged as “Orphan alpha”.
Orphan chains are most likley the effect of stochastic dropouts due to sequencing inefficiencies.
See also
scirpy.tl.chain_pairing().- UMI
Unique molecular identifier. Some single-cell RNA-seq protocols label each RNA with a unique barcode prior to PCR-amplification to mitigate PCR bias. With these protocols, UMI-counts replace the read-counts generally used with RNA-seq.
- productive chain
Productive chains are TCR chains with a CDR3 sequence that produces a functional peptide. Scirpy relies on the preprocessing tools (e.g. CellRanger or TraCeR) for flagging non-productive chains. Typically chains are flagged as non-productive if they contain a stop codon or are not within the reading frame.