API
Import scirpy together with scanpy as
import scanpy as sc
import scirpy as ir
For consistency, the scirpy API tries to follow the scanpy API as closely as possible.
Input/Output: io
Note
In scirpy v0.7.0 the way VDJ data is stored in adata.obs has changed to
be fully compliant with the AIRR Rearrangement
schema. Please use upgrade_schema() to make AnnData objects
from previous scirpy versions compatible with the most recent scirpy workflow.
|
Update older versions of a scirpy anndata object to the latest schema. |
The following functions allow to import V(D)J information from various formats.
|
Read |
|
Read IR data from 10x Genomics cell-ranger output. |
|
Read data from TraCeR ([SLonnbergP+16]). |
|
|
|
Read IR data from the BD Rhapsody Analysis Pipeline. |
|
Read data from AIRR rearrangement format. |
|
Scirpy can export data to the following formats:
|
|
|
To convert own formats into the scirpy Data structure, we recommend building
a list of AirrCell objects first, and then converting them into
an AnnData object using from_airr_cells().
For more details, check the Data loading tutorial.
|
Data structure for a Cell with immune receptors. |
|
|
|
Convert an adata object with IR information back to a list of |
Preprocessing: pp
|
Merge adaptive immune receptor (IR) data with transcriptomics data into a single |
|
Merge two AnnData objects with IR information (e.g. |
|
Computes a sequence-distance metric between all unique VJ CDR3 sequences and between all unique VDJ CDR3 sequences. |
Tools: tl
Tools add an interpretable annotation to the AnnData object
which usually can be visualized by a corresponding plotting function.
Generic
|
Summarizes the number/fraction of cells of a certain category by a certain group. |
Quality control
|
Perform quality control based on the receptor-chain pairing configuration. |
Define and visualize clonotypes
|
Define clonotypes based on CDR3 nucleic acid sequence identity. |
|
Define clonotype clusters. |
|
Finds evidence for Convergent evolution of clonotypes. |
|
Computes the layout of the clonotype network. |
|
Get an |
Analyse clonal diversity
|
Adds a column to |
|
Summarizes clonal expansion by a grouping variable. |
|
Computes the alpha diversity of clonotypes within a group. |
|
Compute distance between cell groups based on clonotype overlap. |
|
Identifies clonotypes or clonotype clusters consisting of cells that are more transcriptionally related than expected by chance by computing the Clonotype modularity. |
|
Query reference databases
|
Query a referece database for matching immune cell receptors. |
|
Annotate cells based on the result of |
|
Returns the inner join of |
V(D)J gene usage
|
Summarizes the distribution of CDR3 region lengths. |
Plotting: pl
Generic
|
A customized wrapper to the |
Tools
Every of these plotting functions has a corresponding tool in the scirpy.tl
section. Depending on the computational load, tools are either invoked on-the-fly
when calling the plotting function or need to be precomputed and stored in
AnnData previously.
|
Plot the alpha diversity per group. |
|
Visualize clonal expansion. |
|
Plots the number of cells per group, split up by a categorical variable. |
|
Show the distribution of CDR3 region lengths. |
|
Creates a ribbon plot of the most abundant VDJ combinations. |
|
Visualizes overlap betwen a pair of samples on a scatter plot or |
|
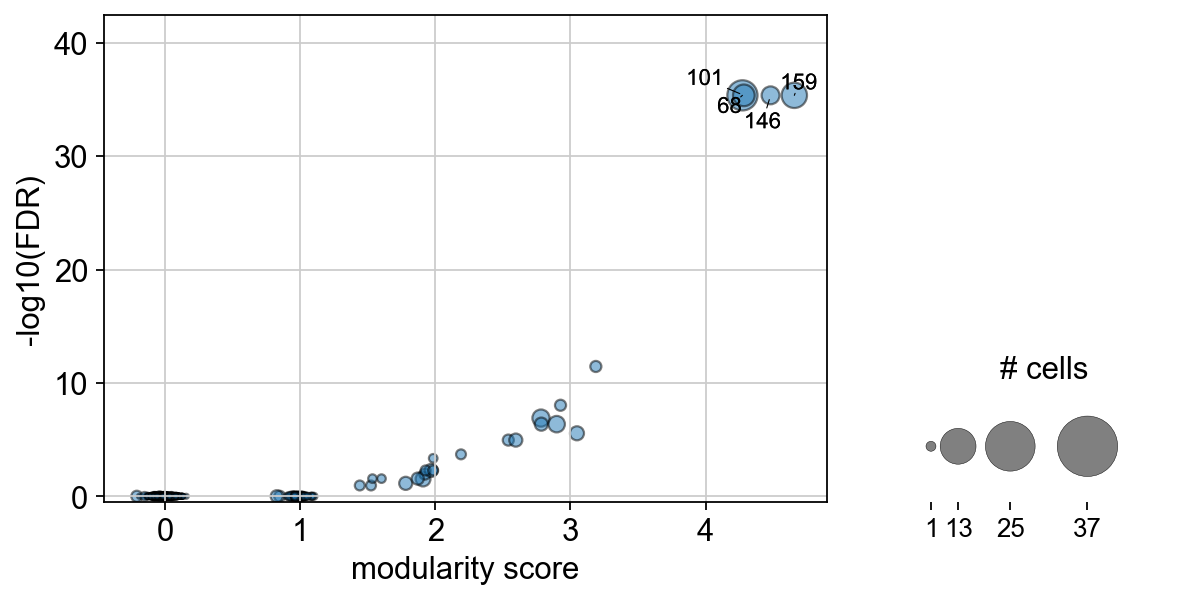
Plots the Clonotype modularity score against the associated log10 p-value. |
|
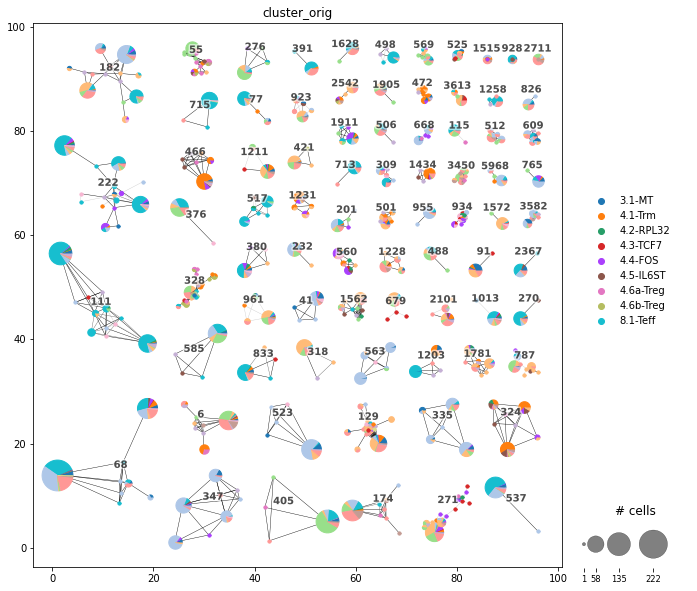
Plot the Clonotype network. |
|
Aims to find clonotypes that are the most enriched or depleted in a category. |
Base plotting functions: pl.base
|
Basic plotting function built on top of bar plot in Pandas. |
|
Basic plotting function built on top of line plot in Pandas. |
|
Basic plotting function built on top of bar plot in Pandas. |
|
Basic plotting function for drawing KDE-smoothed curves. |
Plot styling: pl.styling
|
Apply a predefined style to an axis object. |
|
Style an axes object. |
Datasets: datasets
Return the dataset from [WMdA+20] as AnnData object. |
|
Return the dataset from [WMdA+20] as AnnData object, downsampled to 3000 TCR-containing cells. |
|
Return the dataset from [MMR+20] as AnnData object. |
Reference databases
|
Download VDJdb and process it into an AnnData object. |
|
Download IEBD v3 and process it into an AnnData object. |
A reference database is also just a Scirpy-formatted AnnData object. This means you can follow the instructions in the data loading tutorial to build a custom reference database.
Utility functions: util
|
Compute a graph layout by layouting all connected components individually. |
|
Compute the Fruchterman-Reingold layout respecting node sizes. |
|
Get an igraph object from an adjacency or distance matrix. |
IR distance utilities: ir_dist
|
Calculate a sequence x sequence distance matrix. |
distance metrics
Abstract base class for a CDR3-sequence distance calculator. |
|
Abstract base class for a DistanceCalculator that computes distances in parallel. |
|
Calculates the Identity-distance between CDR3 sequences. |
|
Calculates the Levenshtein edit-distance between sequences. |
|
Calculates the Hamming distance between sequences of identical length. |
|
Calculates distance between sequences based on pairwise sequence alignment. |